My learnings from ML4Science @IIITH

“I believe there is no more important application for AI than helping to improve human health.”
~ Demis Hassabis
Inspired by this assessment of AI use case, and with a fundamental background in machine learning, I decided to venture into Scientific ML, one that not only excites me but I believe I can bring significant impact with it to the world.
In line with this, I recently got a privileged opportunity to attend ML4Science Winter School at International Institute of Information Technology Hyderabad (IIITH) under the guidance of Prof Deva Priyakumar, that primarily aimed at bridging the scientific research community to machine learning.
This article is not intended to provide the understanding of the fundamentals but rather aims at giving the reader a glimpse of the concepts taught, the hands-on experiments we worked on, a go-through of the project I worked on during the cohort as well as provide an effective refresher whenever I come back to it.
Day-wise Progression of the Cohort
Day 1:
- Prof Deva Priyakumar began with an introductory session to the ML4Science, what to expect from the course and built a headspace wrapped around the topics to come.
Day 2:
- Both the lectures on day 2 by Prof CV Jawahar laid the perfect foundations of machine learning and neural networks to build on from here (No one could have done it better than him, it was like learning batting from Bradman, simply the top in the domain)
- We learned core machine learning concepts, overfitting vs. generalization, and neural networks: from simple linear classifiers to deep networks, including loss optimization, forward/backpropagation, and matrix-based computation.
- We focused on practical work using Python ML libraries, built neural networks in PyTorch, extracted molecular features with RDKit, and applied decision tree and MLP models to chemical regression and classification tasks (ESol dataset).
Day 3:
-
The first lecture of the day was conducted by Prof Deva Priyakumar, where we were introduced to the concept of Representation of Molecules for its use in ML model including SMILES notation, Coulomb Matrix, Fingerprint representations.
-
The second lecture was conducted by Prof Maitreya Maity and Prof Vinod P K on subsequent topics in deep learning i.e, CNNs and RNNs and their application in scientific domain. We got a detailed overview of CNN concepts including Convolutions, Kernels, Pooling and how does it help to capture the spatial information.
-
This was followed by an explanation of RNN: how can model built using recurrent neurons help in capturing temporal dependence and variants of RNNs (LSTM, Bi-LSTM, GRU, Bi-GRU, Vanilla and Bidirectional RNN)
-
During our hands-on, we trained a Molecular CNN model, aimed at demonstrating how deep learning can directly operate on molecular geometry rather than hand-crafted features, as required by traditional ML models. We converted a chemical molecular structure into 3D voxel grids using CNN based on SMILES notation, which then learns spatial and chemical patterns from these 3D representations to predict molecular properties.
-
We also worked on MLP and Decision tree based models tasks using MACCS fingerprints, Morgan (ECFP) Fingerprints, Coulomb matrix, ACSF Descriptors. The primary task was to predict a property of a molecule (aqueous solubility, partition coefficient) using different represenation methods.
Day 4:
-
After CNN (spatial modeling) and RNN (sequential modeling), we learnt about models used for describing and analyzing entities with interactions, using Graph Representation Learning (GRL) taken by Prof. Charu Sharma, focusing on the concept of Knowledge graphs, Graph Neural Networks (GNN), categories of tasks (node, edge, subgraph, and graph based), methods of GRL, as well as applications of graph in scientific discipline: finding similar protein motifs and drug repurposing.
-
We followed GRL with Unsupervised Learning taught by Dr. Monalisa Patra, diving deep into building models where no explicit target is given. We learnt about multiple approaches in an Unsupervised learning problem, including Density Estimation using Parametric (Gaussian mixture model) and Non Parametric Estimation methods, Dimensionality Reduction (PCA, Multi-Dimensional Scaling & tSNE for visualisation of high dimensional data), Clustering (similarity and density based) and Detection of Anomalies. What defined this lecture was that we were given a fundamental understanding of different approaches for a given problem based on the structure of data at hand.
-
The practicals of the day focused on building sequential models to capture the structure of protein sequence: built LSTM, RNN, Bi-directional RNN and Bi-directional GRU and classified their secondary protein structure from given amino acid sequence.
-
We also practically applied three distinct Graph based models: Graph attention Network (GAT), Graph Convolutional Network (GCN), and a vanilla Graph Neural Network (GNN) to predict aqueous solubility, partition coefficient of a given molecule.
Day 5:
-
Day 5 was one of the most exciting days as it included discussion on topics “Generative Model” and “Transformers” : the current talk of the town.
The lecture for Generative Models was conducted by Prof Girish Verma, where we began with understanding the taxonomy or classification of the generative models into Explicit Density and Implicit Density models and their further discussion. We also explored generations of generative models starting from models based on normalizing flow, autoencoders, variational autoencoders (VAE), generative adversarial networks (GAN). -
The second lecture of the day taken by Prof Vineet Gandhi introduced the core concept of text based generation models ie, Transformer Models, starting from as fundamental as tokens to reaching attention mechanism of the transformer model. We understood different types of attention mechanisms, including self-attention, multi-head attention, masked attention mechanisms and positional encodings. This lecture fundamentally helped us build an understanding of the engine that runs the modern day LLMs. In addition, we understood the scientific application of these architectures: Protein Language Models (AlphaFold, ESM), transformer based SMILES to Property generator of a molecule, something that I later worked on during my project.
-
The practical sessions of the day involved building following models: Autoencoder, VAE, Conditional and Unconditional GPT as well as a transformer to learn the sequence based structure of proteins. We worked on autoencoders to discover its dimensionality reduction feature wherein we trained a model to reduce a high-dimensionality fingerprint of a molecule into its lower dimension latent space still capable to retain key properties of the molecule.
-
We trained a VAE model that can generate SMILE strings for novel molecular structures, using an encoder-latent space-decoder architecture, where the model encodes molecular structures into a continuous latent space, decode latent vectors back into valid molecular structures to generate new molecules by sampling from this learned latent space.
-
In a addition we trained two classes of GPT based generative models to generate valid molecules (SMILES representation) by predicting one character at a time, similar to text generation. The Unconditional GPT generates molecules randomly based on learned patterns with no control over molecular properties, whereas the conditional model generates molecules with specific target properties (logP, SAS), like a molecule with logP=2.5 and SAS=3.0. Such specific generation is popular in generating molecules/drugs with target properties. The only difference between the two architectures being an addition of a property embedding network corresponding to the desired properties.
Day 6:
-
Moving on from text generation models, day 6 involved exploring the modern image generation model architecture, ie, Diffusion model. The lecture was conducted by Prof Monalisa Patra, who explained the complete architecture of the Markov chain based diffusion model: Forward Diffusion, where small random noise (perturbations) are added to the data AND Backward Diffusion, where the model learns small changes to go from noise to data, basically trying to calculate how much noise was added at every step. We further learnt about specific architectures of Diffusion Models, including U-Net, Latent Space based Diffusion model and Diffusion Transformer.
-
The second lecture of the day was conducted by Prof Girish Verma, covering conceptual understanding of Reinforcement Learning. We were familiarized with idea of independent decision making amongst agents, through the State-Action-Reward (+ Transition probability) concept based from Markov’s Decision Process. The explanation was further enriched with concepts of Policy, State-Value Function and encompassing Deep Q-learning.
-
Similar to the practicals for GPT, the hands-on session for Diffusion model involved building conditional and unconditional models to generate either diverse or target specific molecules. Apart from text/image generation, the major difference was between the GPT and diffusion models was the use of SELFIES molecular representation instead of SMILES.
-
The second practical of the day, the last of the winter school, involved training model based on Reinforcement Learning, where the agent is a GPT based Conditional SMILES Generator model, with POLICY to optimize for the target LogP value of the molecule. The task was to fine-tune a pre-trained conditional GPT model for the target-directed molecular generation. This is a betterment with respect to the previous generative approach which are handicapped to learn from data distributions as RL helps to optimize the generator model directly for specific objectives using reward signals.
Day 7:
- The final day of the cohort was the D-Day where we had to present our projects to the panel. I will be talking more about my project in the next section. Prior to these presentations, we were given an exciting tour of the Agentic AI landscape by Prof Karthik Vaidhyanathan, who with his captivating demonstration introduced us to the capabilities of AI when given an autonomy of execution. We understood that behind every AI-System is a “system of multiple systems”, and that each has to perform optimally to provide the desired outcome.
Most rewarding part of the cohort: PROJECT WORK

The most rewarding and enriching part of my time during this cohort was when I worked on an inter-disciplinary project, wherein our team had to build a Hemolytic Concentration (HC50) prediction model, given ONLY the peptide sequence.
During my research work, I found out that HC50 is a component of Eco-toxicity: a critical measurement of the harmful impact of a molecule, or a drug, on an unintended entity. Therefore pharmaceutical eco-toxicity assessment becomes a crucial regulatory requirement, which in turn makes correct predictions of eco-toxicity measures absolutely indispensable.
The Opportunity:
The systems or algorithms that predict the eco-toxicity levels are known as QSAR models. The regulatory bodies accept QSAR results only if they comply with the multiple validation principles. Since most of the QSAR are either structure based, similarity based or property descriptor based, there is an opportunity to use Deep Learning based models, especially sequential models, to develop a modern QSAR.
The advantage of DL based models will be: after adequate pre-training, and use-case based fine-tuning, these QSAR models will be relatively faster to work with, thus reducing testing costs as well as enables the testing of a drug or newly synthesized compound at multiple stages. They will also help in automatically extracting complex features, handling non-linear relationships, and processing diverse chemical datasets better than traditional methods.
 Source: ditki medical & biological sciences
Source: ditki medical & biological sciences
The Challenge:
As evident the order of the amino acids in protein determine the structure and properties of the protein, it was CRITICAL to preserve this sequence.
To enrich the feature vector, we need to derive appropriate features from the external sources for each sequence.
The Models Experimented With:
During the course of this project we tried multiple approaches to find the best performing, optimized model, evaluated on the metrics : Root Mean Squared Error and Spearman's Coefficient.
A. Bidirectional LSTM + XGBoost
- Focusing on primarily the first challenge ie "preserving the sequence", I decided to try an ensemble of a traditional sequential model coupled with a regression model, in this case XGBoost
B. Using feature extraction library : Pfeature
- Pfeature is a feature-generation toolkit developed at IIIT-Delhi that converts raw peptide/protein sequences into machine-learning–ready descriptors.
- Using Pfeature, we derived Molecular weight, Amino Acid Composition, Dipeptide Composition, Hydrophobicity, Net Charge among others, based on the bio-related disciplinary understanding.
- As our regression, we used an ensemble model coupling random forest and extra trees regressor.
C. Using Protein Language Model (PLM): BEST PERFORMING
- The breakthrough moment of the project was when we discovered the concept of PLM (a model that learns patterns in protein sequences to understand their structure and function), introduced by Prof Vinod.
- We used Meta's ESM2 (650M parameters) model to act as feature extractor to create embeddings that were then fed into the downstream regression model (Random Forest in this case)
The Outcome:
The model we built gave satisfactory baseline levels of performance with an RMSE of 0.4914 (for the log scale HC50 value), and Spearman's Coefficient of 0.6462.
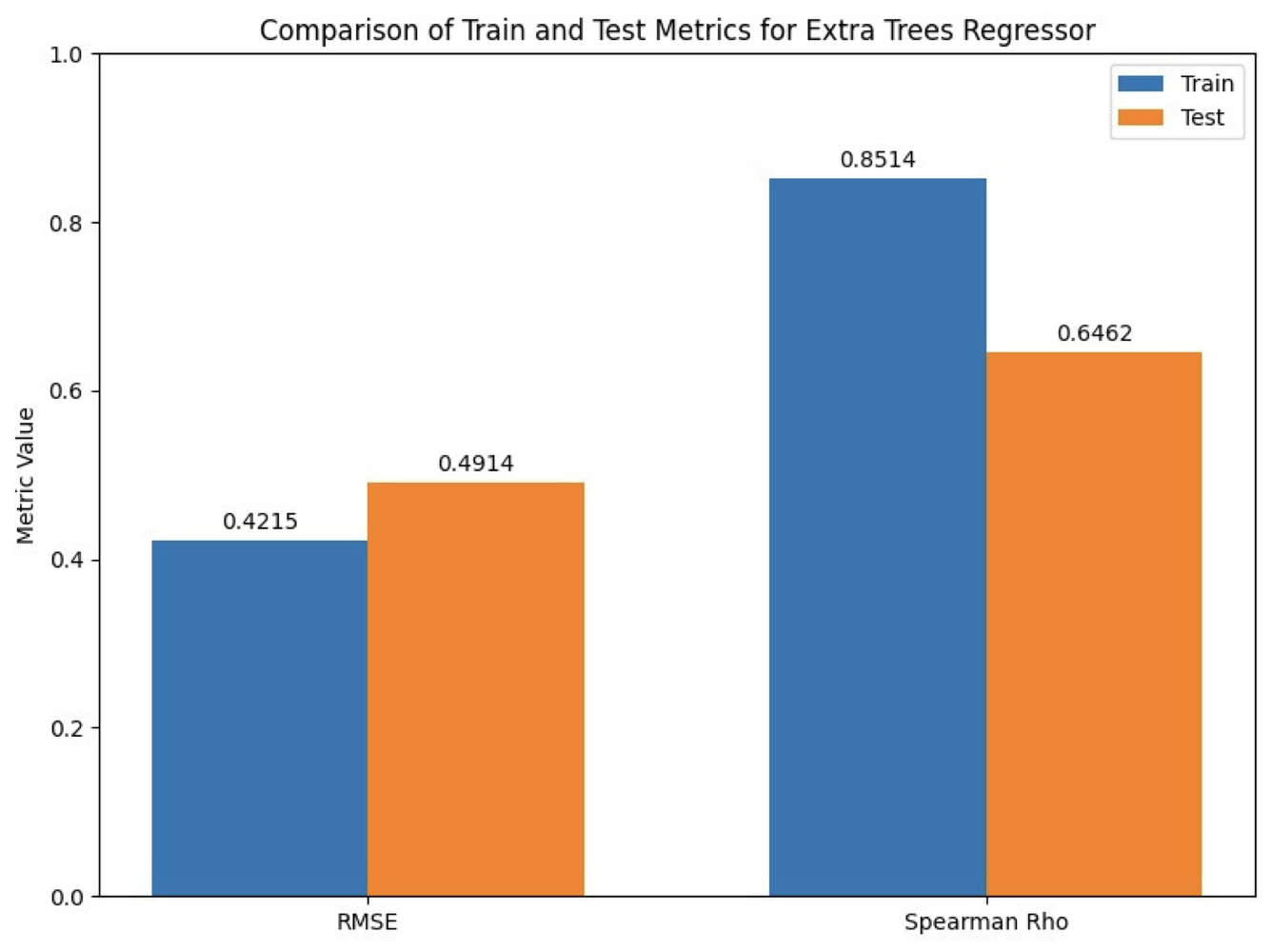 In the graph: Final model metrics
In the graph: Final model metrics
Parting remarks
AI's generative capacity has largely been successful in capturing the intrigue and imagination of the industries across domains, however the most effective application of AI will be enhancing the process of scientific discovery, due to its speed and ability to learn from intricacies of the data.
During the course of this winter school, I had the opportunity to connect and hear about the ongoing projects of multiple PhD scholars from top research institutes of India including Indian Institute of Technology, Bombay, Indian Institute of Science Education and Research (IISER), Pune, Indian Institute of Science (IISc), among others. These meetings helped me find out the direction of scientific research and the gaps to apply ML in.
I'm eager to apply the knowledge built and experience gained to projects, research and applications that bring real impact to the society. Looking for opportunities to work with labs, researchers and industry-experts to leverage machine learning & deep learning in achieving scientific breakthroughs.
Way to go,
Aryan